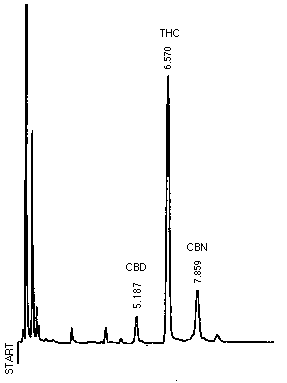
Fig.1 Chromatogram of GC
CBD :Cannabidiol
THC :Tetrahydrocannabinol
CBN :Cannabinol
Syuji OKUYAMA and Toshiyuki MITSUI
 Return
Return
Table 1 Operating conditions of GC/MS
------------------------------------------------
Instrument JEOL JMS-DX300
MS-GCG05
Column 1.5% Silicone OV-17
(2.5mm i.d. x 1m)
Column temp. 230C
Injection temp. 250C
Separator temp. 250C
Inlet temp. 250C
Chamber temp. 200C
Ionization volt 70V
Ionization current 300.micro.A
Carrier gas He
------------------------------------------------
Table 2 Operating conditions of GC
--------------------------------------------------
Insturument HEWLETT PACKARD
5890 SERIES II
Column DB1 (0.53mm i.d. x 15m)
Column temp. 230C
Injection temp. 250C
Detecter temp. 250C
Carrier gas He
-------------------------------------------------

Fig.1 Chromatogram of GC
CBD :Cannabidiol
THC :Tetrahydrocannabinol
CBN :Cannabinol
those of the selected peaks and fragment ions, they were hardly differentiated among marihuana and were not useful in the cluster analysis. In these selected peaks and fragment ions, THC for GC and 314 in THC for GC-MS were used as the internal standards. For GC, the areas of CBD and CBN were divided by the area of THC. For GC-MS, the area of selected fragment ions was divided by the area of 314 in THC. With combining the selected peaks and fragment ions of GC and GC-MS, respectively, 9 values were obtained from each sample. According to Table 3, the values were then normalized to eleven blocks from 0 to 10. Because the intact values divided by the internal standard included experimental errors, this normalization method
Table 3 Correction of the data
------------------------------------------------
Divided value by Normalized
internal standard(%) number
------------------------------------------------
0 0
0 - 1 1
1 - 2.5 2
2.5- 5 3
5 - 7.5 4
7.5- 10 5
10 - 25 6
25 - 50 7
50 - 75 8
75 -100 9
100 - 10
------------------------------------------------
gave smaller errors than those detected when the values divided by the internal standard were used. An example of these calculations is shown in Table 4. The corrected values are shown in Table 5. The cluster analysis was performed using this matrix.
Table 4 Normalization method of peak area
----------------------------------------------
Peak Peak Divided Normalized
number area value by I.S. value by Table3
----------------------------------------------
I.S. 494.8 --- ---
1 517.4 1.046 10
2 0 0 0
3 243.2 0.492 7
4 0 0 0
5 0 0 0
6 379.2 0.766 9
7 21.4 0.043 3
I.S. 12438 --- ---
8 485 0.039 3
9 63 0.005 1
----------------------------------------------
Table 5 Filed data for cluster analysis
--------------------------------------------------------------------
Sample number Peak number Sample number Peak number
--------------------------------------------------------------------
1 2 3 4 5 6 7 8 9 1 2 3 4 5 6 7 8 9
1 10 0 7 3 0 9 3 6 2 23 10 2 8 2 3 9 5 6 1
2 10 0 7 0 0 9 3 3 1 24 9 3 7 3 0 9 6 5 3
3 10 2 8 2 0 9 3 2 1 25 10 3 8 3 0 9 4 3 6
4 10 2 8 2 3 9 6 6 8 26 10 2 8 3 0 9 2 5 6
5 10 0 8 0 2 9 2 2 6 27 9 1 8 2 0 10 2 6 2
6 9 0 8 0 0 9 3 3 3 28 9 1 8 1 4 9 6 4 6
7 10 0 8 0 6 8 10 10 10 29 9 2 8 3 2 10 7 7 4
8 10 5 8 4 0 9 4 3 2 30 10 2 8 2 2 9 6 5 6
9 10 0 8 0 2 9 5 2 8 31 10 1 8 2 3 9 7 6 6
10 10 0 8 4 7 9 8 10 10 32 10 2 8 2 6 9 7 6 7
11 9 3 7 3 2 9 4 5 6 33 10 6 8 6 0 9 6 3 6
12 10 0 8 0 0 9 3 3 1 34 10 6 8 6 0 9 6 4 6
13 10 0 8 0 0 9 4 3 1 35 10 5 8 4 0 9 5 3 4
14 9 0 8 0 0 9 2 3 1 36 10 3 8 3 2 9 4 6 4
15 10 2 8 2 2 9 7 6 1 37 10 2 8 2 2 7 3 6 1
16 10 2 8 2 3 9 7 6 1 38 10 0 8 0 0 9 2 2 6
17 10 2 8 2 3 9 8 6 1 39 10 0 8 0 0 9 2 8 6
18 10 5 8 5 0 9 3 3 6 40 10 4 8 4 0 9 4 5 3
19 10 3 8 3 0 9 3 3 5 41 10 0 8 0 0 9 2 3 6
20 10 3 8 3 0 9 3 3 6 42 9 2 8 1 0 10 0 6 6
21 10 6 8 6 0 9 3 3 6 43 9 2 8 1 3 9 5 6 3
22 10 4 8 3 0 9 3 3 4
--------------------------------------------------------------------
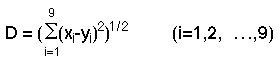
The MIED indicates the similarity of samples. We judged whether the samples
were the same or not using the MIED. The judgment was determined from an experiment
consisting of more than five hundred samples. If the MIED was less than five, we
judged these samples as the same. If it was larger than five and less than
ten, these samples were similar. Further, if it was larger than ten and
less than twenty, those samples might be similar. A dendrogram was
prepared with the use of the MIED and is shown in
Figure 3. This
presents an easily interpreted visual representation of the similarity among
samples. Two samples that were connected more closely were more similar than the other.
In this dendrogram, we judged that samples 16 and 17 belong to the same
group. Similarly, samples 12 and 13, 19 and 20, 33 and 34 were judged to belong
to the same group. Actually, these are the samples that were seized from the same
suspect and the Euclidean distances are less than five. Samples 19 and 25 were
seized from different suspects but these belong to the same group because the
MIED between two samples is less than five. It turned out that these samples
were purchased from the same seller.

Fig.3 Dendrogram of cluster analysis
 Return
Return