CyberMol - A Molecular Graphics Program System on
the World Wide Web Using the Common Gateway Interface
Hiroshi YOSHIDA and Hiroatsu MATSUURA
 Return
Return
1. INTRODUCTION
The explosive development of the
World Wide Web (WWW) has changed the computer network dramatically.
Programs on the computer connected to the Internet can be easily
operated by the WWW browser using the Common Gateway Interface
(CGI). Furthermore, three-dimensional (3D) graphics on the Internet
has recently been developed and its new technique, the Virtual
Reality Modeling Language (VRML), presents the visualization of
molecular models by using the VRML viewer. The 3D technique based
on the VRML can be applied effectively in chemistry [1-3].
In the present work, we have developed
a 3D molecular graphics program system, CyberMol, which
can visualize, by using the CGI, molecular models on the WWW by
the use of the VRML viewer. This system also provides the organic
molecular structure database and the search engine for displaying
molecular models by using the VRML viewer.
2. IMPLEMENTATION
We use an IBM/AT compatible (DOS/V)
machine, in which the Microsoft Windows NT 3.51 Workstation operating
system has been installed. The EMWAC
HTTP Server for Windows NT (version 0.991) program
[4] is used for an HTTP server. The home page of the CyberMol,
whose URL is
http://vbl01.chem.sci.hiroshima-u.ac.jp/CyberMol1.0/,
is written in the Hypertext Markup Language (HTML). A Sun workstation
at the Information Processing Center of Hiroshima University is
also used for executing the CGI scripts written in the Perl language.
These scripts generate a VRML format text according to the corresponding
inputted molecular coordinates and transport it to the HTTP server.
The DOS/V machine and the Sun workstation are connected to the
Internet and can be accessed to use the CyberMol. We tested the
CyberMol by using a Netscape Navigator which plugged-in a Live3D
as a VRML viewer.
3. OVERVIEW OF CYBERMOL AND DISCUSSION
When the home page of the CyberMol
is accessed through the Internet, all of the menus are displayed
on the WWW browser as shown in Fig. 1. Selection of a menu opens
the corresponding page. The function and the usage of the relevant
pages are given below.
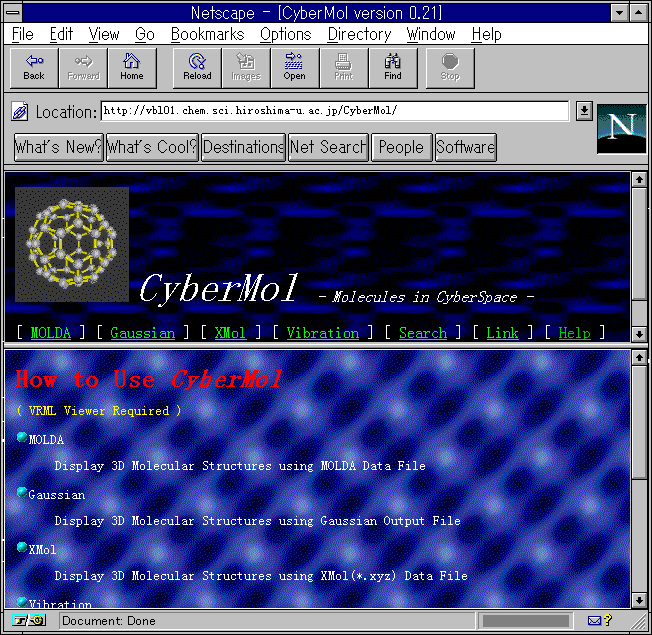
Fig. 1 The home page of the CyberMol.
MOLDA
The 'MOLDA'
page deals with the display of 3D molecular models using MOLDA
format data [3,5]. The molecular coordinates and the type of molecular
models (Dreiding sticks, ball-and-stick and spacefilling models
are available) are inputted as shown in Fig. 2. By pushing a 'Go!'
button, the molecular model is displayed as shown in Fig. 3.
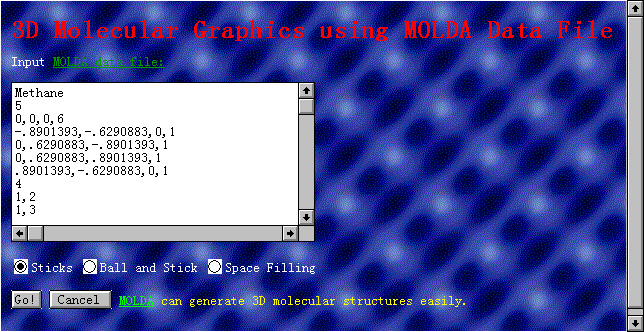
Fig. 2 The 'MOLDA' page.
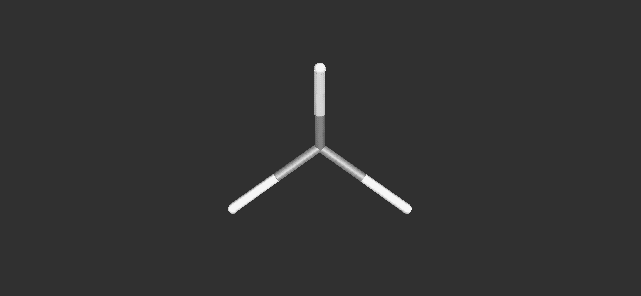
Fig. 3 A Dreiding stick model of methane.
Gaussian
The 'Gaussian'
page deals with the display of 3D molecular models using the GAUSSIAN
94 output file [6]. The operation is the same as that of the 'MOLDA'
page.
XMol
The 'XMol'
page deals with the display of 3D molecular models using XMol
format data (*.xyz) [7]. The operation is the same as that of
the 'MOLDA' page.
Vibration
The 'Vibration'
page deals with the visualization of vibrational modes using the
GAUSSIAN 94 output file. The molecular coordinates and the displacements
of atoms calculated by the GAUSSIAN 94 program are inputted as
shown in Fig. 4. By pushing a 'Go!'
button, the vibrational mode is displayed as shown in Fig. 5.
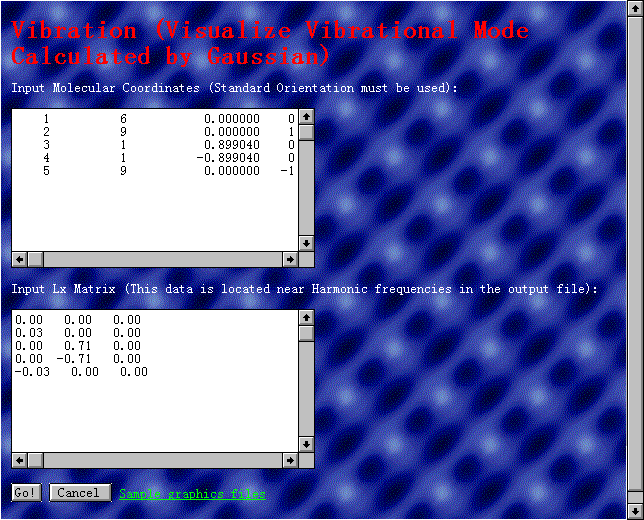
Fig. 4 The 'Vibration' page.
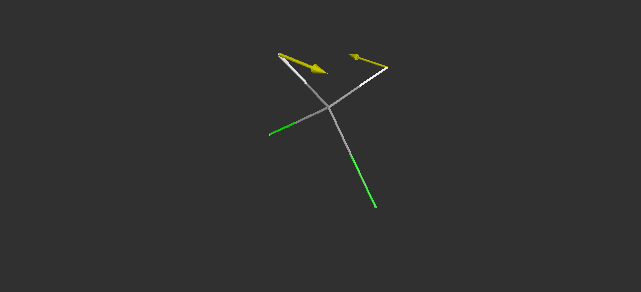
Fig. 5 The CH2 twisting vibration of CH2F2.
Search
The 'Search'
page deals with the search of molecular coordinates from the organic
molecular structure database provided by Nagao [8]. To search
for the molecular structure data, enter a keyword (a Perl regular
expression is also available). After searching, the ball-and-stick
model of the molecule is displayed on the VRML viewer.
Link
The 'Link'
page deals with the links to other VRML chemistry resources.
Help
The 'Help'
page deals with the help for using the CyberMol.
By using these functions, the CyberMol
can be easily operated by the WWW browser using the CGI not only
by local users but also by remote users who can use terminal computers
which are connected to the Internet. Moreover, the system is used
platform-independently; in other words, the molecular models can
be displayed on the DOS/V, Macintosh and Unix machines by using
the CyberMol. For a long time, computer software has been dependent
on hardware and operating systems. On the contrary, the present
system provides a new programming paradigm that software should
work platform-independently. The new programming approach will
change the programming styles in computer chemistry.
4. CONCLUSION
CyberMol provides molecular graphics
on the WWW by using the MOLDA, XMol and GAUSSIAN 94 format data.
The organic molecular structure database and the search engine
for displaying molecular models by using the VRML viewer are useful
in chemical education. The system can be easily operated by the
WWW browser using the CGI not only by local users but also by
remote users who can use terminal computers which are connected
to the Internet. In the next stage, the CyberMol will offer the
function of the molecular modeling and control the ab initio molecular
orbital calculation and molecular dynamics simulation programs
by the combined use of the CGI, VRML and Java technologies. This
forthcoming system will generate the new age of molecular science
and molecular technology.
REFERENCES
1) H. Vollhardt, G. Moeckel, C.
Henn, M. Teschner and J. Brickmann, "VRML
for the communication with 3D scenarios of biomolecules,"
http://ws05.pc.chemie.th-darmstadt.de/vrml/.
2) O. Casher and H. S. Rzepa, "Chemical
Examples of VRML,"
http://www.ch.ic.ac.uk/VRML/.
3) H. Yoshida, "MOLDA
Home Page,"
http://vbl01.chem.sci.hiroshima-u.ac.jp/molda/; H. Yoshida and H. Matsuura, J. Chem. Software, 3, 147 (1997)
4) The European Microsoft Windows
NT Academic Centre, "Freeware
HTTP Server for Windows NT,"
http://emwac.ed.ac.uk/html/internet_toolchest/https/contents.htm.
5) K. Ogawa, H. Yoshida and H. Suzuki,
J. Mol. Graphics, 2, 113 (1984).
6) M. J. Frisch, G. W. Trucks, H.
B. Schlegel, P. M. W. Gill, B. G. Johnson, M. A. Robb, J. R. Cheeseman,
T. Keith, G. A. Petersson, J. A. Montgomery, K. Raghavachari,
M. A. Al-Laham, V. G. Zakrzewski, J. V. Ortiz, J. B. Foresman,
J. Cioslowski, B. B. Stefanov, A. Nanayakkara, M. Challacombe,
C. Y. Peng, P. Y. Ayala, W. Chen, M. W. Wong, J. L. Andres, E.
S. Replogle, R. Gomperts, R. L. Martin, D. J. Fox, J. S. Binkley,
D. J. Defrees, J. Baker, J. P. Stewart, M. Head-Gordon, C. Gonzalez
and J. A. Pople, Gaussian 94, Revision D.3, Gaussian, Inc., Pittsburgh,
PA, 1995.
7) Minnesota Supercomputer Center,
Inc., "XMol (Version1.5)
User Guide,"
http://www.kfajuelich.de/zam/CompServ/software/graph/xmol/XMol.html.
8) T. Nagao, CSSJ Software, Registry
No. 9104.
 Return
Return

 Return
Return
 Return
Return